Overview
The NYU Center for Genomics and Systems Biology (CGSB) Genomics Core (Gencore) bioinformatics team delivers high-throughput sequencing data with speed, automation, and reliability. Researchers get end-to-end automation, expert support for nonstandard library designs, and consistently fast turnaround across platforms. Built for scale and low friction, our computational platform accelerates discovery and sets a high bar for flexibility, engineering, and service.
Beyond data processing, we advance the research and education mission of NYU through hands-on training, purpose-built tools, tailored bioinformatics support, and active course/student engagement, ensuring investigators at every stage can fully leverage our advanced computational resources.
The Team
Sequencing Platforms We Support
Gencore supports multiple technologies to meet the diverse needs of the NYU research community:
Illumina: MiSeq, NextSeq, NovaSeq
Oxford Nanopore (ONT): MinIOn and GridION
Element Biosciences: AVITI
To simplify and streamline workflow, researchers use a single computational system to submit metadata for sequencing, track progress, and access results across all sequencing platforms.
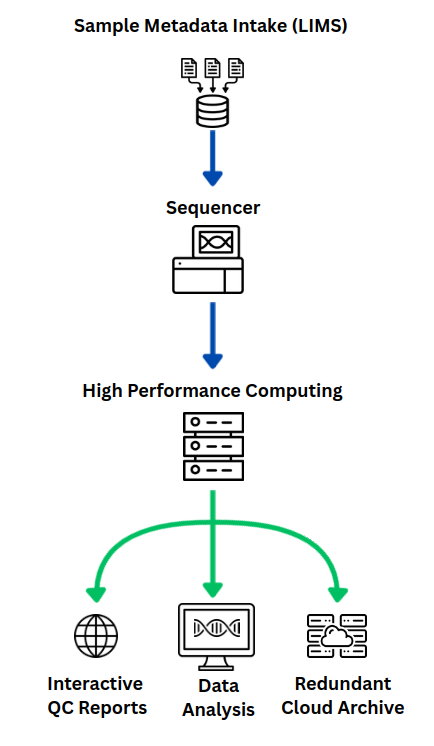
How We Manage, Process, and Deliver Your Data
Our fully automated custom analysis pipeline handles every step of sequencing data processing: from basecalling and demultiplexing to quality control and archiving. Built with open-source tools and engineered to scale efficiently across NYU’s HPC cluster, it is designed for speed, flexibility, and resilience.
We support advanced and custom sequencing library designs, automatically detect and correct common technical issues, and integrate with our Laboratory Information Management System (LIMS), TuboWeb, which tracks all sample metadata and processing parameters. Because all metadata and processing inputs are recorded at the point of sample submission, the system ensures every result is traceable and reproducible.
Upon completion, researchers receive interactive Quality Control (QC) reports alongside clean, structured data—delivered via the NYU HPC cluster (accessible via terminal or web portal) or direct transfer to external collaborators. We also support processing runs sequenced at other centers, offering the same streamlined delivery and reliability. All datasets are permanently, and redundantly, archived and can be restored on demand, so labs never have to worry about managing or losing sequencing data.
Integration with NYU HPC
Our computational bioinformatics infrastructure is fully integrated with the NYU HPC cluster. This collaboration is pivotal to the success of our sequencing data workflows and overall research capability.
We install and maintain required bioinformatics software, manage storage, and enforce secure, permissioned access.
With Open OnDemand, researchers use a web interface to manage and download data; launch GPU-accelerated Jupyter Notebooks, RStudio, and IGV; and run command-line tasks—all within a user-friendly graphical environment.
Hardware upgrades occur behind the scenes; software and pipelines remain available throughout, so transitions to new infrastructure are seamless and research continues without disruption.
Tools, Resources, and Infrastructure
We regularly assist researchers with selecting the right tools, optimizing their workflows, and resolving technical challenges across the entire analysis lifecycle. Whether navigating complex pipeline frameworks, troubleshooting errors and resolving failures, addressing large-scale data handling challenges, or managing compute resources, we provide hands-on guidance that empowers researchers to move forward with confidence.
We support a rich ecosystem of tools, pipelines, and resources that empower researchers to analyze, visualize, and share their data. All Gencore developed software and pipelines are maintained at: https://github.com/gencorefacility/
JBrowse: Web-based genome browser. Upload custom datasets, share views publicly, and link directly to published results.
NASQAR: Web-based genomics suite for RNA-seq, metagenomics, epigenetics, variant calling, and more—modular, easy, and built for single-cell and bulk data.
reform and reform WebApp: A fast, robust, and flexible Python tool for editing reference genome sequence and annotation files.
GATK4 Variant Calling Pipeline: A fast, scalable, and portable pipeline optimized for the NYU HPC, includes built-in QC, and outputs directly to JBrowse for interactive review.
RShiny Server: Hosts custom-built, interactive applications tied to user datasets—ideal for sharing and publishing.
Shared Genome Resource: A curated collection of commonly used genome references and indexes, maintained on NYU HPC.
Next-Generation Sequencing Educational Portal: Our genomics educational portal features guides, tutorials, and walkthroughs for common tools and analysis workflows, attracting 10,000+ monthly visitors at NYU and globally.
MinION Basecalling on GPU: GPU-accelerated basecalling for Oxford Nanopore on NYU HPC, enabling high-throughput processing for labs running ONT locally.
Custom Bioinformatics Solutions: We work with researchers to develop tailored bioinformatics solutions.
Nextflow + nf-core Support: Reproducible, open-source, community-driven pipelines optimized for the NYU HPC.
Bioinformatics and Data Analysis Seminars (BADAS): Gencore’s flagship seminar series offers researchers at all levels exposure to new bioinformatics tools, techniques, and best practices.