Gaining Access to Sequencing Data on the HPC
- Create an HPC account by following the directions found here: NYU High Performance Computing Wiki.
- Submit a request via the Biology Computation Support Form to be added to the CGSB Linux working group on the HPC, and to be granted permission to your lab’s sequencing results directory.
Data Policy and Retention
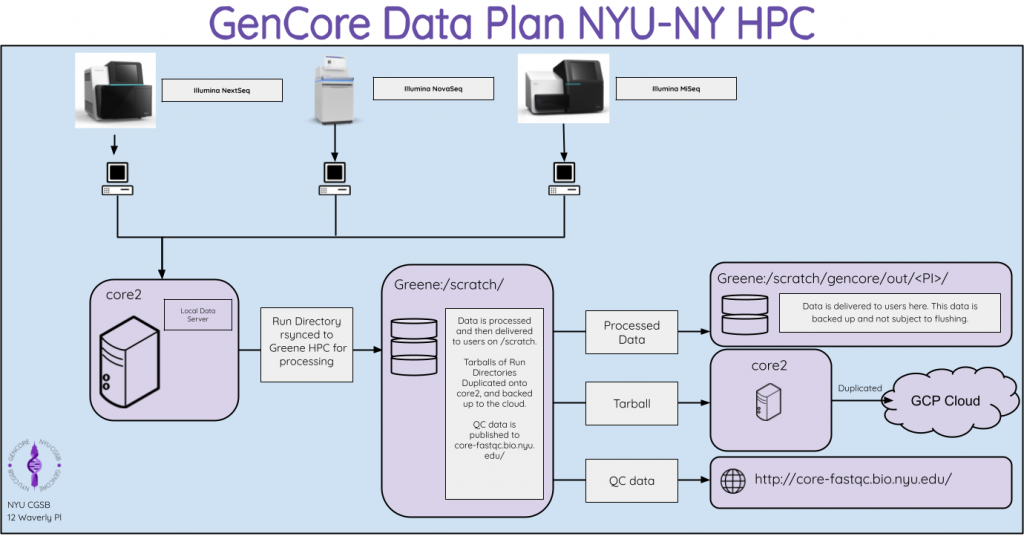
- Demultiplexed fastqs or raw lane fastqs are copied to lab directories on
/scratchon the HPC. - Owners of the data will have read access to the fastqs on
/scratch. - Sequencing data in lab directories on
/scratchare backed up and not subject to flushing. - Raw and processed sequencing run directories are archived and backed up locally for a minimum of five years.
- Raw sequencing run directories can be made available to users on request.
- Lab shares are kept up to 3 years after PI departure from CGSB.
HPC Best Practices
- Your job should run in, and output written to, your personal directory on scratch. i.e.
/scratch/netID/my-project/job-xyz/ - Your Slurm scripts should live in your job or project directory
- Keeping a copy of the job script in it’s run directory is good practice as it allows you to check later what parameters were used and facilitates reproducibility.
- All other scripts (ex: python scripts, other executables) should live in your home folder (ie.
/home/netID)/ - If you need to run a script that you created (a python script for example), call it from your home directory (accessible via the $HOME variable) in your slurm script
- If you need a software package which is not available on the HPC, please email the HPC team at hpc@nyu.edu with your request. You can check if a module already exists by typing
module avail tool_nameon the command line.
HPC Important Locations
Fastq Delivery from GenCore
/scratch/cgsb/gencore/out/
- Files here are not subject to flushing
- Files here are backed up
Lab Share Directory
/scratch/cgsb/
- Data stored here is subject to storage charges
- Files here are not subject to flushing
- Files here are backed up to the cloud
- Use this directory to share your analysis and results with other members of your lab
- Reference and input data (ex: GenCore delivered fastq files, reference genome, indexes, etc.) should not live here!
- To establish a labshare directory on /scratch/cgsb and to add members to your Labshare directory submit a request using this form
Personal Directory on scratch
/scratch/netID/
- 5TB Quota
- Files here which are not used for a period of 60 days are subject to flushing.
- Use this directory to run your analysis and store analysis results as you are working on them
Personal Directory on home
/home/netID/
- 50GB Quota
- Not subject to flushing
- Backed up
- Your custom scripts (python scripts or other executables) should live here
Personal Directory on archive
/archive/netID/
- 2TB Quota
- Not subject to flushing
- Data will be backed up
- Completed analyses and results that you want to store should be archived (tar) and then stored here
Shared Genome Resource
/scratch/work/cgsb/genomes
- Local CGSB repository of commonly used genomic data sets
- New organisms/versions/releases will be made available periodically or upon request (Mohammed Khalfan – mkhalfan@nyu.edu)
- Previous versions/releases will be preserved
- More about the Shared Genome Resource…
Recent Articles
Nextflow & nf-core on NYU HPC
All nextflow and nf-core pipelines have been successfully configured for use on the HPC Cluster at New York University. The configuration applies required and recommended...
Non fungible tokens (NFTs) for academic publications?
Being rejected from the preprint server, bioRxiv, seemed like a new low for me. But, I was heartened to know that I was in good...
Streamlined RNA-Seq Analysis Using Nextflow
UPDATED: April 16, 2024 nf-core is a community effort to collect a curated set of analysis pipelines built using Nextflow. This post will walk you...