Bioinformatics
Variant Calling Pipeline using GATK4
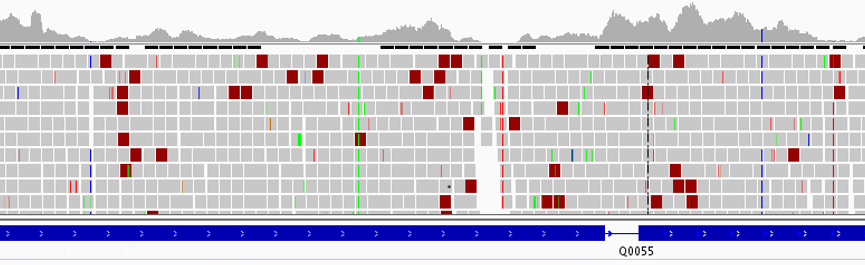
This is an updated version of the variant calling pipeline post published in 2016 (link). This updated version employs GATK4 and is available as a containerized Nextflow script on GitHub. Identifying genomic variants, including single nucleotide polymorphisms (SNPs) and DNA insertions and deletions (indels), from next generation sequencing data is Read more