The bioinformatics team at the NYU Center for Genomics and Systems Biology in Abu Dhabi and New York have recently developed NASQAR (Nucleic Acid SeQuence Analysis Resource), a web-based platform providing an intuitive interface to popular R-based bioinformatics data analysis and visualization tools including Seurat, DESeq2, Shaman, clusterProfiler, and more.
These tools, although powerful, typically require significant computational experience and lack a graphical user interface (GUI), making them inaccessible to many researchers. NASQAR addresses this problem by wrapping these tools in a beautifully simple UI, allowing users to explore their data in seconds using various tools and visualization methods. Users also have the ability to download analysis results and a wide array of figures.
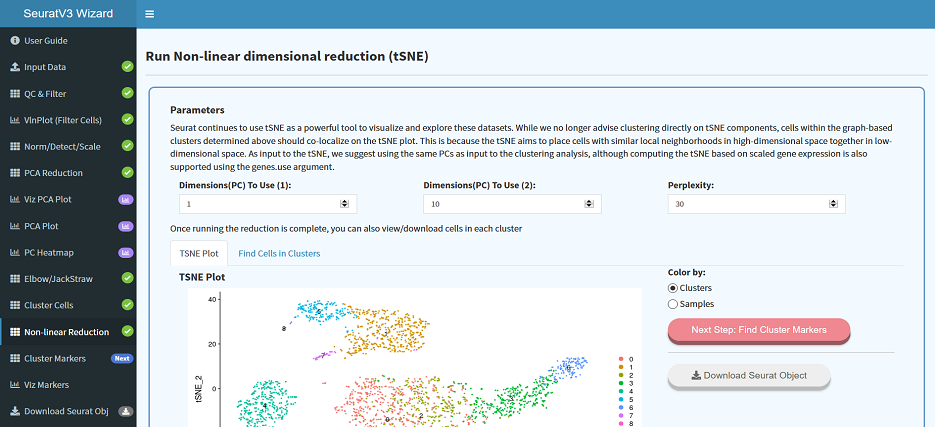
Behind the scenes data pre-processing allows for a seamless transition to downstream analysis, and sample data included with each app allows users to take a test drive without having any data on hand.
The web app lowers the entry barrier and provides greater independence for researchers with little or no computational experience to carry out standard bioinformatics analysis, visualize their data, and produce publication-ready data files and figures.
The platform is publicly accessible at http://nasqar.abudhabi.nyu.edu/.
NASQAR is open-source and the code is available through GitHub at https://github.com/nasqar/NASQAR.
NASQAR is also available as a Docker image at https://hub.docker.com/r/aymanm/nasqarall.
Let us know what you think in the comments below!

0 Comments